Pneumocystis carinii EST (cDNA) Analysis
The goal of the Pneumocystis Expressed Sequence Tag portion of the project is to provide a rapid gene inventory of genes expressed in Pneumocystis.
Background
Two libraries were analyzed to initiate the Pneumocystis Expressed Sequence Tag portion of the project: a well-known library produced by Jeff Edman and a new library constructed by George Smulian. The Edman library contains multiple short inserts and significant host contamination that prevent its use. The Smulian library contains long inserts and host contamination of 14%, an acceptable level for this purpose. We sequenced ~4500 clones from the Smulian library.
Details of library construction. The library was constructed from RNA isolated from a single rat with a Form 1 P. carinii infection (See karyotype on this page). The library was generated in lambdaZAPII (Stratagene) cloned directionally between the EcoRI and XhoI sites. It consisted of 5x105 primary clones and contains ~14% host contamination. EST sequence was obtained from an aliquot of this unamplified library rescued with helper phage to yield pBluscript SKII+ plasmids containing insets. The library was amplified once to a titer of 9x1011. Aliquots of this material are available from George Smulian. The average insert size is about 1.5 kb. The majority of the inserts appear to be full length. The cDNAs were sequenced at the University of Georgia. Data analysis continues to be conducted by the University of Cincinnati (College of Medicine / Division of Infectious Diseases) using Children's Hospital Medical Center of Cincinnati (CHMCC) Bioinformatics Computer Systems. The cDNA sequences were screened for vector and contaminants (Rat cDNA) using the Phred and Cross Match genomic processing tools. Then the sequences were futher trimmed, using 2 custom developed perl programs (qualityScoreBasedTrim.pl and qualityScoreBasedFastaSeqExtraction.pl). These programs required each cDNA sequence read to have a miminum of 50 bases with a maximum expected quality score error rate of 1.0 and are available for download from the software portion of this web site. The sequences reads were extended until the maximum error was reached. Next, the sequences were assembled using the Cap3 Assembler to reduce redundancy and increase read reliability by condensing overlapping sequence and associated quality scores.
Pc cDNA Homology Table
We compiled our EST dataset by removing duplicate mRNA sequences using the Cap3 assembly results and BlastN/X homology data using Genbank's NT (Nucleotide) and NR (Amino Acid) databases. In addition, the homology data was employed to identify a set of Unique Pc Genes. The UNIQUE SET of Pc ESTs includes Genbank Homology Names, Descriptions, and Scores and is available for download as an EXCEL table and for browsing on-line.
Pc cDNA Fasta Sequences
Our Unigene Data Set is composed of duplicate read cDNAs (contigs) and single read cDNAs (singlets). By COMBINING the cDNA contigs and singlets, we have a non-redundant set of Unique cDNAs, represented in the above Pc GENES with Genbank Homology Names in an Excel Table. The SINGLET FASTA sequences and the CONTIG FASTA sequences are both available. Please note that the sequence names in the Pc GENES Excel file correlate to these fasta sequences.
 |
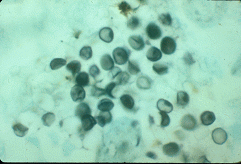 |
| Form 1 P. carinii (left) and P. carinii cysts (top) stained with methanamine silver. |
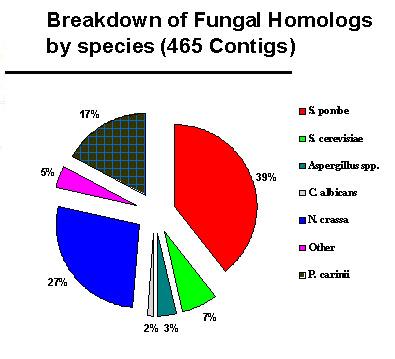
The assembled EST sequences (Contigs) showed most homology to
fungal genes:
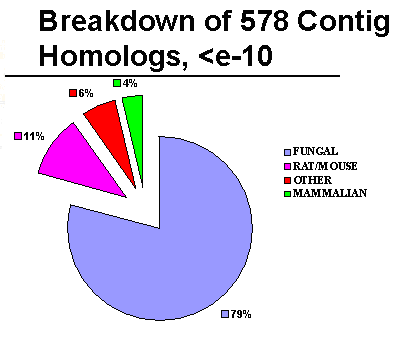
Most of the fungal homologies were to Schizosaccharomyces
pombe and Neurospora crassa:
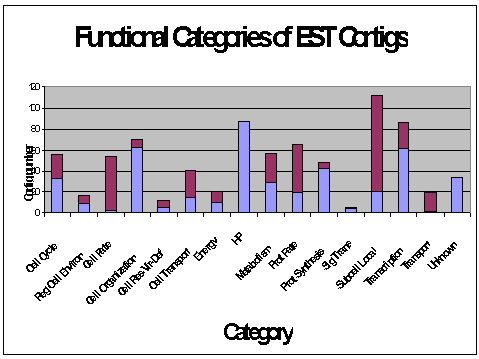
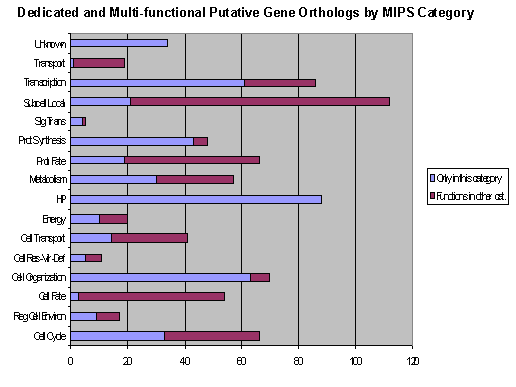
A variety of metabolic cycles and cellular processes were
represented in the ESTs: